Introduction to R - Tidyverse
rpi.analyticsdojo.com
Overview
It is often said that 80% of data analysis is spent on the process of cleaning and preparing the data. (Dasu and Johnson, 2003)
Thus before you can even get to doing any sort of sophisticated analysis or plotting, you’ll generally first need to:
- Manipulating data frames, e.g. filtering, summarizing, and conducting calculations across groups.
- Tidying data into the appropriate format
What is the Tidyverse?
Tidyverse
- “The tidyverse is a set of packages that work in harmony because they share common data representations and API design.” -Hadley Wickham
- The variety of packages include
dplyr,tibble,tidyr,readr,purrr(and more).
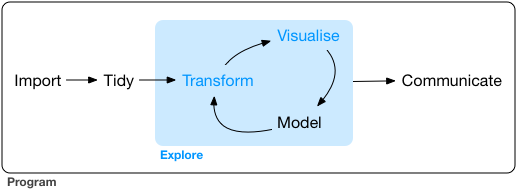
- From R for Data Science by Hadley Wickham
Schools of Thought
There are two competing schools of thought within the R community.
- We should stick to the base R functions to do manipulating and tidying;
tidyverseuses syntax that’s unlike base R and is superfluous. - We should start teaching students to manipulate data using
tidyversetools because they are straightfoward to use, more readable than base R, and speed up the tidying process.
We’ll show you some of the tidyverse tools so you can make an informed decision about whether you want to use base R or these newfangled packages.
Dataframe Manipulation using Base R Functions
- So far, you’ve seen the basics of manipulating data frames, e.g. subsetting, merging, and basic calculations.
- For instance, we can use base R functions to calculate summary statistics across groups of observations,
- e.g. the mean GDP per capita within each region:
gapminder <- read.csv("../../input/gapminder-FiveYearData.csv",
stringsAsFactors = TRUE)
head(gapminder)
But this isn’t ideal because it involves a fair bit of repetition. Repeating yourself will cost you time, both now and later, and potentially introduce some nasty bugs.
Dataframe Manipulation using dplyr
Here we’re going to cover 6 of the most commonly used functions as well as using pipes (%>%) to combine them.
select()filter()group_by()summarize()mutate()arrange()
If you have have not installed this package earlier, please do so now:
install.packages('dplyr')
Dataframe Manipulation using dplyr
Luckily, the dplyr package provides a number of very useful functions for manipulating dataframes. These functions will save you time by reducing repetition. As an added bonus, you might even find the dplyr grammar easier to read.
- “A fast, consistent tool for working with data frame like objects, both in memory and out of memory.”
- Subset observations using their value with
filter(). - Reorder rows using
arrange(). - Select columns using
select(). - Recode variables useing
mutate(). - Sumarize variables using
summarise().
#Now lets load some packages:
library(dplyr)
library(ggplot2)
library(tidyverse)
dplyr select
Imagine that we just received the gapminder dataset, but are only interested in a few variables in it. We could use the select() function to keep only the columns corresponding to variables we select.
year_country_gdp <-gapminder[,c("year","country")]
year_country_gdp
year_country_gdp <- select(gapminder, year, country, gdpPercap)
head(year_country_gdp)
dplyr Piping
%>%Is used to help to write cleaner code.- It is loaded by default when running the
tidyverse, but it comes from themagrittrpackage. - Input from one command is piped to another without saving directly in memory with an intermediate throwaway variable. -Since the pipe grammar is unlike anything we’ve seen in R before, let’s repeat what we’ve done above using pipes.
year_country_gdp <- gapminder %>% select(year,country,gdpPercap)
dplyr filter
Now let’s say we’re only interested in African countries. We can combine select and filter to select only the observations where continent is Africa.
As with last time, first we pass the gapminder dataframe to the filter() function, then we pass the filtered version of the gapminder dataframe to the select() function.
To clarify, both the select and filter functions subsets the data frame. The difference is that select extracts certain columns, while filter extracts certain rows.
Note: The order of operations is very important in this case. If we used ‘select’ first, filter would not be able to find the variable continent since we would have removed it in the previous step.
year_country_gdp_africa <- gapminder %>%
filter(continent == "Africa") %>%
select(year,country,gdpPercap)
dplyr Calculations Across Groups
A common task you’ll encounter when working with data is running calculations on different groups within the data. For instance, what if we wanted to calculate the mean GDP per capita for each continent?
In base R, you would have to run the mean() function for each subset of data:
mean(gapminder[gapminder$continent == "Africa", "gdpPercap"])
mean(gapminder[gapminder$continent == "Americas", "gdpPercap"])
mean(gapminder[gapminder$continent == "Asia", "gdpPercap"])
dplyr split-apply-combine
The abstract problem we’re encountering here is know as “split-apply-combine”:

We want to split our data into groups (in this case continents), apply some calculations on each group, then combine the results together afterwards.
Module 4 gave some ways to do split-apply-combine type stuff using the apply family of functions, but those are error prone and messy.
Luckily, dplyr offers a much cleaner, straight-forward solution to this problem.
# remove this column -- there are two easy ways!
dplyr group_by
We’ve already seen how filter() can help us select observations that meet certain criteria (in the above: continent == "Europe"). More helpful, however, is the group_by() function, which will essentially use every unique criteria that we could have used in filter().
A grouped_df can be thought of as a list where each item in the list is a data.frame which contains only the rows that correspond to the a particular value continent (at least in the example above).

#Summarize returns a dataframe.
gdp_bycontinents <- gapminder %>%
group_by(continent) %>%
summarize(mean_gdpPercap = mean(gdpPercap))
head(gdp_bycontinents)

That allowed us to calculate the mean gdpPercap for each continent. But it gets even better – the function group_by() allows us to group by multiple variables. Let’s group by year and continent.
gdp_bycontinents_byyear <- gapminder %>%
group_by(continent, year) %>%
summarize(mean_gdpPercap = mean(gdpPercap))
gdp_bycontinents_byyear
mpg<-mpg
str(mpg)
That is already quite powerful, but it gets even better! You’re not limited to defining 1 new variable in summarize().
gdp_pop_bycontinents_byyear <- gapminder %>%
group_by(continent, year) %>%
summarize(mean_gdpPercap = mean(gdpPercap),
sd_gdpPercap = sd(gdpPercap),
mean_pop = mean(pop),
sd_pop = sd(pop))
head(gdp_pop_bycontinents_byyear)
Basics
- Use the mpg dataset to create summaries by manufacturer/year for 8 cyl vehicles.
mpg<-mpg
head(mpg)
#This just gives a dataframe with 70 obs, only 8 cylinder cars
mpg.8cyl<-mpg %>%
filter(cyl == 8)
mpg.8cyl
#Filter to only those cars that have miles per gallon equal to
mpg.8cyl<-mpg %>%
filter(cyl == 8)
#Alt Syntax
mpg.8cyl<-filter(mpg, cyl == 8)
mpg.8cyl
#Sort cars by MPG highway(hwy) then city(cty)
mpgsort<-arrange(mpg, hwy, cty)
mpgsort
#From the documentation https://cran.r-project.org/web/packages/dplyr/dplyr.pdf
select(iris, starts_with("petal")) #returns columns that start with "Petal"
select(iris, ends_with("width")) #returns columns that start with "Width"
select(iris, contains("etal"))
select(iris, matches(".t."))
select(iris, Petal.Length, Petal.Width)
vars <- c("Petal.Length", "Petal.Width")
select(iris, one_of(vars))
#Recoding Data
# See Creating new variables with mutate and ifelse:
# https://rstudio-pubs-static.s3.amazonaws.com/116317_e6922e81e72e4e3f83995485ce686c14.html
mutate(mpg, displ_l = displ / 61.0237)
# Example taken from David Ranzolin
# https://rstudio-pubs-static.s3.amazonaws.com/116317_e6922e81e72e4e3f83995485ce686c14.html#/9
section <- c("MATH111", "MATH111", "ENG111")
grade <- c(78, 93, 56)
student <- c("David", "Kristina", "Mycroft")
gradebook <- data.frame(section, grade, student)
#As the output is a tibble, here we are saving each intermediate version.
gradebook2<-mutate(gradebook, Pass.Fail = ifelse(grade > 60, "Pass", "Fail"))
gradebook3<-mutate(gradebook2, letter = ifelse(grade %in% 60:69, "D",
ifelse(grade %in% 70:79, "C",
ifelse(grade %in% 80:89, "B",
ifelse(grade %in% 90:99, "A", "F")))))
gradebook3
#Here we are using piping to do this more effectively.
gradebook4<-gradebook %>%
mutate(Pass.Fail = ifelse(grade > 60, "Pass", "Fail")) %>%
mutate(letter = ifelse(grade %in% 60:69, "D",
ifelse(grade %in% 70:79, "C",
ifelse(grade %in% 80:89, "B",
ifelse(grade %in% 90:99, "A", "F")))))
gradebook4
#find the average city and highway mpg
summarise(mpg, mean(cty), mean(hwy))
#find the average city and highway mpg by cylander
summarise(group_by(mpg, cyl), mean(cty), mean(hwy))
summarise(group_by(mtcars, cyl), m = mean(disp), sd = sd(disp))
# With data frames, you can create and immediately use summaries
by_cyl <- mtcars %>% group_by(cyl)
by_cyl %>% summarise(a = n(), b = a + 1)
#This was adopted from the Berkley R Bootcamp.
