Coronavirus Data Modeling
Background
From Wikipedia…
“The 2019–20 coronavirus pandemic is an ongoing global pandemic of coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The virus was first reported in Wuhan, Hubei, China, in December 2019.[5][6] On March 11, 2020, the World Health Organization declared the outbreak a pandemic.[7] As of March 12, 2020, over 134,000 cases have been confirmed in more than 120 countries and territories, with major outbreaks in mainland China, Italy, South Korea, and Iran.[3] Around 5,000 people, with about 3200 from China, have died from the disease. More than 69,000 have recovered.[4]
The virus spreads between people in a way similar to influenza, via respiratory droplets from coughing.[8][9][10] The time between exposure and symptom onset is typically five days, but may range from two to fourteen days.[10][11] Symptoms are most often fever, cough, and shortness of breath.[10][11] Complications may include pneumonia and acute respiratory distress syndrome. There is currently no vaccine or specific antiviral treatment, but research is ongoing. Efforts are aimed at managing symptoms and supportive therapy. Recommended preventive measures include handwashing, maintaining distance from other people (particularly those who are sick), and monitoring and self-isolation for fourteen days for people who suspect they are infected.[9][10][12]
Public health responses around the world have included travel restrictions, quarantines, curfews, event cancellations, and school closures. They have included the quarantine of all of Italy and the Chinese province of Hubei; various curfew measures in China and South Korea;[13][14][15] screening methods at airports and train stations;[16] and travel advisories regarding regions with community transmission.[17][18][19][20] Schools have closed nationwide in 22 countries or locally in 17 countries, affecting more than 370 million students.[21]”
https://en.wikipedia.org/wiki/2019–20_coronavirus_pandemic
For ADDITIONAL BACKGROUND, see JHU’s COVID-19 Resource Center: https://coronavirus.jhu.edu/
#RPI IDEA
Check out these resources that IDEA has put together.
https://idea.rpi.edu/covid-19-resources
The Assignment
Our lives have been seriously disrupted by the coronavirus pandemic, and there is every indication that this is going to be a global event which requires colloration in a global community to solve. Studying the data provides an opportunity to connect the pandemic to the variety of themes from the class.
A number of folks have already been examining this data. https://ourworldindata.org/coronavirus-source-data
- Discussion. What is the role of open data? Why is it important in this case?
```###Answer here. q1=”””
”””
</div>
</div>
2. Read this.
https://medium.com/@tomaspueyo/coronavirus-act-today-or-people-will-die-f4d3d9cd99ca
What is the role of bias in the data? Identify 2 different ways that the data could be biased.
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```###Answer here.
q2="""
"""
```#Load some data import pandas as pd df=pd.read_csv(‘https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_daily_reports/03-22-2020.csv’) df
</div>
<div class="output_wrapper" markdown="1">
<div class="output_subarea" markdown="1">
<div markdown="0" class="output output_html">
<div>
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
</style>
<table border="1" class="dataframe">
<thead>
<tr style="text-align: right;">
<th></th>
<th>Province/State</th>
<th>Country/Region</th>
<th>Last Update</th>
<th>Confirmed</th>
<th>Deaths</th>
<th>Recovered</th>
<th>Latitude</th>
<th>Longitude</th>
</tr>
</thead>
<tbody>
<tr>
<th>0</th>
<td>Hubei</td>
<td>China</td>
<td>2020-03-22T09:43:06</td>
<td>67800</td>
<td>3144</td>
<td>59433</td>
<td>30.9756</td>
<td>112.2707</td>
</tr>
<tr>
<th>1</th>
<td>NaN</td>
<td>Italy</td>
<td>2020-03-22T18:13:20</td>
<td>59138</td>
<td>5476</td>
<td>7024</td>
<td>41.8719</td>
<td>12.5674</td>
</tr>
<tr>
<th>2</th>
<td>NaN</td>
<td>Spain</td>
<td>2020-03-22T23:13:18</td>
<td>28768</td>
<td>1772</td>
<td>2575</td>
<td>40.4637</td>
<td>-3.7492</td>
</tr>
<tr>
<th>3</th>
<td>NaN</td>
<td>Germany</td>
<td>2020-03-22T23:43:02</td>
<td>24873</td>
<td>94</td>
<td>266</td>
<td>51.1657</td>
<td>10.4515</td>
</tr>
<tr>
<th>4</th>
<td>NaN</td>
<td>Iran</td>
<td>2020-03-22T14:13:06</td>
<td>21638</td>
<td>1685</td>
<td>7931</td>
<td>32.4279</td>
<td>53.6880</td>
</tr>
<tr>
<th>...</th>
<td>...</td>
<td>...</td>
<td>...</td>
<td>...</td>
<td>...</td>
<td>...</td>
<td>...</td>
<td>...</td>
</tr>
<tr>
<th>304</th>
<td>NaN</td>
<td>Jersey</td>
<td>2020-03-17T18:33:03</td>
<td>0</td>
<td>0</td>
<td>0</td>
<td>49.1900</td>
<td>-2.1100</td>
</tr>
<tr>
<th>305</th>
<td>NaN</td>
<td>Puerto Rico</td>
<td>2020-03-22T22:43:02</td>
<td>0</td>
<td>1</td>
<td>0</td>
<td>18.2000</td>
<td>-66.5000</td>
</tr>
<tr>
<th>306</th>
<td>NaN</td>
<td>Republic of the Congo</td>
<td>2020-03-17T21:33:03</td>
<td>0</td>
<td>0</td>
<td>0</td>
<td>-1.4400</td>
<td>15.5560</td>
</tr>
<tr>
<th>307</th>
<td>NaN</td>
<td>The Bahamas</td>
<td>2020-03-19T12:13:38</td>
<td>0</td>
<td>0</td>
<td>0</td>
<td>24.2500</td>
<td>-76.0000</td>
</tr>
<tr>
<th>308</th>
<td>NaN</td>
<td>The Gambia</td>
<td>2020-03-18T14:13:56</td>
<td>0</td>
<td>0</td>
<td>0</td>
<td>13.4667</td>
<td>-16.6000</td>
</tr>
</tbody>
</table>
<p>309 rows × 8 columns</p>
</div>
</div>
</div>
</div>
</div>
### Preprocessing
We have to deal with missing values first.
First let's check the missing values for each column.
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```df.isnull().sum()
Data Reporting
#TBD
For the last update value, we could create a feature that as equal to the number of days since the last report. We might eliminate data that is too old.
```#TBD For the last update value, we could create a feature that as equal to the number of
</div>
</div>
### Missing Values and data
3. How might we deal with missing values? How is the data structured such that aggregation might be relevant.
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```###Answer here.
q3="""
"""
```#Note the country is then the index here. country=pd.pivot_table(df, values=[‘Confirmed’, ‘Deaths’, ‘Recovered’], index=’Country/Region’, aggfunc=’sum’)
</div>
</div>
### Clustering
Here is and example of the elbow method, which is used to understand the number of clusters.
https://scikit-learn.org/stable/modules/clustering.html#k-means
The K-means algorithm aims to choose centroids that minimise the inertia, or within-cluster sum-of-squares criterion.
By looking at the total inertia at different numbers of clusters, we can get an idea of the appropriate number of clusters.
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```#This indicates the
from sklearn.cluster import KMeans
sum_sq = {}
for k in range(1,30):
kmeans = KMeans(n_clusters = k).fit(country)
# Inertia: Sum of distances of samples to their closest cluster center
sum_sq[k] = kmeans.inertia_
```#ineria at different levels of K sum_sq
</div>
<div class="output_wrapper" markdown="1">
<div class="output_subarea" markdown="1">
{:.output_data_text}
{1: 18388732383.6612, 2: 5781628112.668509, 3: 1437012534.1559324, 4: 437453272.5568181, 5: 249173713.07080925, 6: 133720143.75294116, 7: 101140484.75294116, 8: 70970124.11542442, 9: 41676542.30886076, 10: 30017447.30886076, 11: 16760158.796828683, 12: 8796949.343951093, 13: 4947365.970771144, 14: 3797381.970771144, 15: 2651536.5041044774, 16: 1877813.419029374, 17: 1363350.3819298667, 18: 1024544.2380769933, 19: 722546.6442815806, 20: 643241.4546326294, 21: 509133.6414066776, 22: 418223.49140667764, 23: 317512.6517577265, 24: 261568.8043432082, 25: 216271.43865631748, 26: 184034.2586798829, 27: 153780.61576464615, 28: 127928.16630164167, 29: 109987.48092580127}
</div>
</div>
</div>
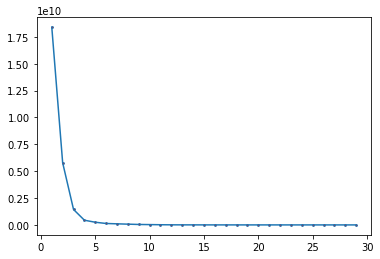
## The Elbow Method
Not a type of criteria like p<0.05, but the elbow method you look for where the change in the variance explained from adding more clusters drops extensively.
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```# plot elbow graph
import matplotlib
from matplotlib import pyplot as plt
plt.plot(list(sum_sq.keys()),
list(sum_sq.values()),
linestyle = '-',
marker = 'H',
markersize = 2,
markerfacecolor = 'red')
#Looks like we can justify 4 clusters.
See how adding the 5th doesn’t really impact the total variance as much? It might be interesting to do the analysis both at 4 and 5 and try to interpret.
```kmeans = KMeans(n_clusters=4) kmeans.fit(country) y_kmeans = kmeans.predict(country)
</div>
</div>
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```y_kmeans
Looks like they are mostly 0s. Let’s merge our data back together so we could get a clearer picture.
```loc=pd.pivot_table(df, values=[‘Latitude’, ‘Longitude’], index=’Country/Region’, aggfunc=’mean’) loc[‘cluster’]=y_kmeans loc
</div>
<div class="output_wrapper" markdown="1">
<div class="output_subarea" markdown="1">
<div markdown="0" class="output output_html">
<div>
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
</style>
<table border="1" class="dataframe">
<thead>
<tr style="text-align: right;">
<th></th>
<th>Latitude</th>
<th>Longitude</th>
<th>cluster</th>
</tr>
<tr>
<th>Country/Region</th>
<th></th>
<th></th>
<th></th>
</tr>
</thead>
<tbody>
<tr>
<th>Afghanistan</th>
<td>33.9391</td>
<td>67.7100</td>
<td>0</td>
</tr>
<tr>
<th>Albania</th>
<td>41.1533</td>
<td>20.1683</td>
<td>0</td>
</tr>
<tr>
<th>Algeria</th>
<td>28.0339</td>
<td>1.6596</td>
<td>0</td>
</tr>
<tr>
<th>Andorra</th>
<td>42.5063</td>
<td>1.5218</td>
<td>0</td>
</tr>
<tr>
<th>Angola</th>
<td>-11.2027</td>
<td>17.8739</td>
<td>0</td>
</tr>
<tr>
<th>...</th>
<td>...</td>
<td>...</td>
<td>...</td>
</tr>
<tr>
<th>Uzbekistan</th>
<td>41.3775</td>
<td>64.5853</td>
<td>0</td>
</tr>
<tr>
<th>Venezuela</th>
<td>6.4238</td>
<td>-66.5897</td>
<td>0</td>
</tr>
<tr>
<th>Vietnam</th>
<td>14.0583</td>
<td>108.2772</td>
<td>0</td>
</tr>
<tr>
<th>Zambia</th>
<td>-13.1339</td>
<td>27.8493</td>
<td>0</td>
</tr>
<tr>
<th>Zimbabwe</th>
<td>-19.0154</td>
<td>29.1549</td>
<td>0</td>
</tr>
</tbody>
</table>
<p>183 rows × 3 columns</p>
</div>
</div>
</div>
</div>
</div>
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```#join in our dataframes
alldata= country.join(loc)
alldata.to_csv("alldata.csv")
alldata
from google.colab import files
files.download("alldata.csv")
```alldata.sort_values(‘cluster’, inplace=True)
</div>
</div>
#How do we interpret our clusters?
<div markdown="1" class="cell code_cell">
<div class="input_area" markdown="1">
```alldata[alldata.cluster!=0]
pd.set_option('display.max_rows', 500) #this allows us to see all rows.
alldata[alldata.cluster==0]
Try some EDA of your own. This is an 10 point in class assignment. LMS (Section 1: In class assignment Clustering) by next Monday 3/30.
Using the Covid-19 Clustering example, try something different as part of the EDA.
Turn in ~1/2 page writeup (NOT A JUPYTER NOTEBOOK) describing what you did.
Examples:
Try visualizing the data differently.
Try running a different clustering algorithm.
Try a different number of clusters. How would it be different if we created ratios that controlled for total population? We tried a different clustering algorithm?